I came across a recent paper by Agarwal and Searls which describes a detailed bibliometric analysis of the scientific literature to identify and characterize specific research topics that appear to be drivers for drug discovery – i.e., research areas/topics in life sciences that exhibit significant activity and thus might be fruitful for drug discovery efforts. The analysis is based on PubMed abstracts, citation data and patents and addresses research topics ranging from very broad WHO disease categories (such as respiratory infectins and skin diseases) to more specific topics via MeSH headings and down to the level of indivudal genes and pathways. The details of the methodology are described in a separate paper, but this one provides some very interesting analyses and visualizations of drug discovery trends.
The authors start out by defining the idea of “push” and “pull”:
Unmet medical need and commercial potential, which are considered the traditional drivers of drug discovery, may be seen as providing ‘pull’ for pharmaceutical discovery efforts. Yet, if a disease area offers no ‘push’ in the form of new scientific opportunities, no amount of pull will lead to new drugs — at least not mechanistically novel ones …
The authors then describe how they characterized “push” and “pull”. The key premise of the paper is that publications rates are an overall measure of activity and when categorized by topic (disease area, target, pathway etc), represent the activity in that area. Of course, there are many factors that characterize why certain therapeutic or target areas receive more interest than others and the authors clearly state that even their concepts of “push” and “pull” likely overlap and thus are not independent.
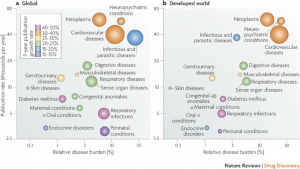
For now I’ll just highlight some of their analysis from the “pull” category. For example, using therapeutic areas from a WHO list (that characterized disease burden) and PubMed searches to count the number of papers in a given area, they generated the plot below. The focus on global disease burden and developed world disease burden was based on the assumption that the former measures general medical need and the latter measures commercial interest.
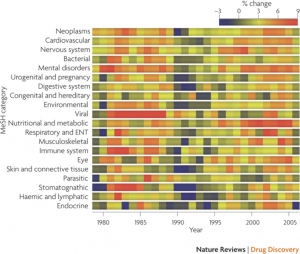
Another interesting summary was the rate of change of publication in a given area (this time defined by MeSH heading). This analysis used a 3 year window over a 30 year period and highlighted some interesting short term trends – such as the spurt in publications for viral research around the early 80’s which likely corresponded to discovery of retroviral involvement in human disease and then later by the AIDS epidemic. The data is summarized in the figure below, where red corresponds to a spurt in activity and blue to a stagnation in publication activity.
They also report analyses that focus on high impact papers, based on a few of the top tier journals (as identified by impact factor) – while their justification is reasonable, it does have downsides which the authors highlight. Their last analysis focuses on specific diseases (based on MeSH disease subcategories) and individual protein targets, comparing the rate of publications in the short term (2 year windows) versus medium term (5 year windows). The idea is that this allows one to identify areas (or targets etc) that exhibit consistent growth (or interest), accelerating and decelerating growth . The resultant plots are quite interesting – though I wonder about the noise involved when going to something as specific as individual targets (identified by looking for gene symbols and synonyms).
While bibliometric analysis is a high level approach the authors make a compelling case that the technique can identify and summarize the drug discovery research landscape. The authors do a great job in rationalizing much of their results. Of course, the analyses are not prospective (i.e., which area should we pour money into next?). I think one of the key features of the work is that it quantitatively characterizes research output and is able to link this to various other efforts (specific funding programs, policy etc) – with the caveat that there are many confounding factors and causal effects.