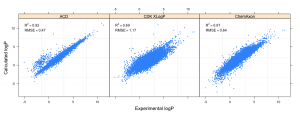
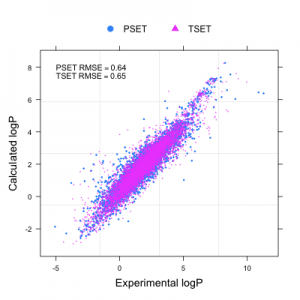
A few weeks back Aaron Sterling posted a review of the Handbook of Cheminformatics Algorithms (in which I have a chapter). Aaron notes
… my goal for the project changed from just a review of a book, to an attempt to build a bridge between theoretical computer science and computational chemistry …
The review/bridging was a pretty thorough summary of the book, but the blog post as well as the comments raised a number of interesting issues that I think are worth discussing. Aaron notes
… Unlike the field of bioinformatics, which enjoys a rich academic literature going back many years, HCA is the first book of its kind …
While the HCA may be the first compilation of cheminformatics-related algorithms in a single place, cheminformatics actually has a pretty long lineage, starting back in the 1960’s. Examples include canonicalization (Morgan, 1965) and ring perception (Hendrickson, 1961). See here for a short history of cheminformatics. Granted these are not CS journals, but that doesn’t mean that cheminformatics is a new field. Bioinformatics also seems to have a similar lineage (see this Biostar thread) with some seminal papers from the 1960’s (Dayhoff et al, 1962). Interestingly, it seems that much of the most-cited literature (alignments etc.) in bioinformatics comes from the 90’s.
Aaron then goes onto note that “there does not appear to be an overarching mathematical theory for any of the application areas considered in HCA“. In some ways this is correct – a number of cheminformatics topics could be considered ad-hoc, rather than grounded in rigorous mathematical proofs. But there are topics, primarily in the graph theoretical areas, that are pretty rigorous. I think Aarons choice of complexity descriptors as an example is not particularly useful – granted it is easy to understand without a background in cheminformatics, but from a practical perspective, complexity descriptors tend to have limited use, synthetic feasibility being one case. (Indeed, there is an ongoing argument about whether topological 2D descriptors are useful and much of the discussion depends on the context). All the points that Aaron notes are correct: induction on small examples, lack of a formal framework for comparison, limited explanation of the utility. Indeed, these comments can be applied to many cheminformatics research reports (cf. “my FANCY-METHOD model performed 5% better on this dataset” style papers).
But this brings me to my main point – many of the real problems addressed by cheminformatics cannot be completely (usefully) abstracted away from the underlying chemistry and biology. Yes, a proof of the lower bounds on the calculation of a molecular complexity descriptor is interesting; maybe it’d get you a paper in a TCS journal. However, it is of no use to a practising chemist in deciding what molecule to make next. The key thing is that one can certainly start with a chemical graph, but in the end it must be tied back to the actual chemical & biological problem. There are certainly examples of this such as the evaluation of bounds on fingerprint similarity (Swamidass & Baldi, 2007). I believe that this stresses the need for real collaborations between TCS, cheminformatics and chemistry.
As another example, Aaron uses the similarity principle (Martin et al, 2002) to explain how cheminformatics measures similarity in different ways and the nature of problems tacked by cheminformatics. One anonymous commenter responds
… I refuse to believe that this is a valid form of research. Yes, it has been mentioned before. The very idea is still outrageous …
In my opinion, the commenter has never worked on real chemical problems, or is of the belief that chemistry can be abstracted into some “pure” framework, divorced from reality. The fact of the matter is that, from a physical point of view, similar molecules do in many cases exhibit similar behaviors. Conversely, there are many cases where similar molecules exhibit significantly different behaviors (Maggiora, 2006). But this is reality and is what cheminformatics must address. In other words, cheminformatics in the absence of chemistry is just symbols on paper.
Aaron, as well as number of commenters, notes that one of the reasons holding back cheminformatics is public access to data and tools. For data, this was indeed the case for a long time. But over the last 10 years or so, a number of large public access databases have become available. While one can certainly argue about the variability in data quality, things are much better than before. In terms of tools, open source cheminformatics tools are also relatively recent, from around 2000 or so. But, as I noted in the comment thread, there is a plethora of open source tools that one can use for most cheminformatics computations, and in some areas are equivalent to commercial implementations.
My last point, which is conjecture on my part, is that one reason for the higher profile of bioinformatics in the CS community is that is has a relatively lower barrier to entry for a non-biologist (and I’ll note that this is likely not a core reason, but a reason nonetheless). After all, the bulk of bioinformatics revolves around strings. Sure there are topics (protein structure etc) that are more physical and I don’t want to go down the semantic road of what is and what is not bioinformatics. But my experience as a faculty member in a department with both cheminformatics and bioinformatics, seems to suggest to me that, coming from a CS or math background, it is easier to get up to speed on the latter than the former. I believe that part of this is due to the fact that while both cheminformatics and bioinformatics are grounded in common, abstract data structures (sequences, graphs etc), one very quickly runs into the nuances of chemical structure in cheminformatics. An alternative way to put it is that much of bioinformatics is based on a single data type – properties of sequences. On the other hand, cheminformatics has multiple data types (aka structural representations) and which one is best for a given task is not always apparent. (Steve Salzberg also made a comment on the higher profile of bioinformatics, which I’ll address in an upcoming post).
In summary, I think Aarons post was very useful as an attempt at bridge building between two communities. Some aspects could have been better articulated – but the fact is, CS topics have been a core part of cheminformatics for a long time and there are ample problems yet to be tackled.
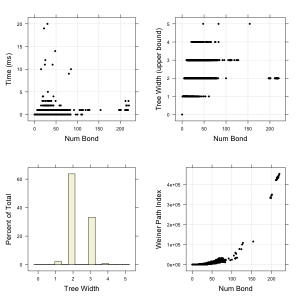 molecules from ChEMBL (also in the Github repository) and evaluated the upper bound of the tree width for each molecule. In addition, I evaluated a few well known topological descriptors for comparison purposes. The four plots summarize the results.
molecules from ChEMBL (also in the Github repository) and evaluated the upper bound of the tree width for each molecule. In addition, I evaluated a few well known topological descriptors for comparison purposes. The four plots summarize the results.