Andrew Dalke recently published a detailed write up on his implementation of the Pubchem fingerprints and provided a pretty thorough comparison with the CDK implementation. He pointed out a number of bugs in the CDK version; but he also noted that performance could be improved by caching parsed SMARTS queries – which are used extensively in this fingerprinter. So I wanted to see whether caching really helps.
The CDK SMARTS parser is a JJTree based implementation and my anecdotal evidence suggested that SMARTS parsing was not a real bottleneck. But of course, nothing beats measurements. So I modified the SMARTSQueryTool class to cache parsed SMARTS queries using a simple LRU cache (patch):
1 2 3 4 5 6 | final int MAX_ENTRIES = 20; Map<String, QueryAtomContainer> cache = new LinkedHashMap<String, QueryAtomContainer>(MAX_ENTRIES + 1, .75F, true) { public boolean removeEldestEntry(Map.Entry eldest) { return size() > MAX_ENTRIES; } }; |
The map is keyed on the SMARTS string. Then, when we attempt to parse a new SMARTS query, we check if it’s in the cache and if not, parse it and place it in the cache.
Now, the thing about this caching mechanism is that after 20 SMARTS queries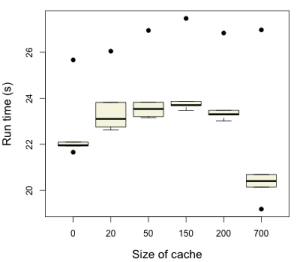 have been cached, the least recently used one is replaced with a new one. As a result, if we perform matching with 40 unique SMARTS (in sequence) only the last 20 get cached, for a given molecule. But when we go to do it on a new molecule, the first 20 are not in the cache and hence we shouldn’t really benefit from the caching scheme. In general, if the fingerprinter (or any caller of the parser) will perform N unique SMARTS queries for a single molecule, the cache size must be at least N, for the cache to be used for subsequent molecules.
have been cached, the least recently used one is replaced with a new one. As a result, if we perform matching with 40 unique SMARTS (in sequence) only the last 20 get cached, for a given molecule. But when we go to do it on a new molecule, the first 20 are not in the cache and hence we shouldn’t really benefit from the caching scheme. In general, if the fingerprinter (or any caller of the parser) will perform N unique SMARTS queries for a single molecule, the cache size must be at least N, for the cache to be used for subsequent molecules.
I implemented a quick test harness, reading in 100 SMILES and then generating Pubchem fingerprints for each molecule. The fingerprint generation was repeated 5 times and the time reported for each round. The box plot shows the distribution of the timings. Now, the CDK implementation has 621 SMARTS patterns – as you can see, we only get a benefit from the caching when the cache size is 700. In fact, cache sizes below that lead to a performance hit – I assume due to time required to query the map.
While the performance improvement is not dramatic it is close to 10% compared to no-caching at all. In actuality, the major bottleneck in the SMARTS parser is the actual graph isomorphism step (which we hope to drastically improve by using the SMSD code). Maybe then, SMARTS parsing will take a bigger fraction of the time. Also keep in mind that due to the heavyweight nature of CDK molecule objects, very large caches could be a strain on memory. But to check that out, I should use a profiler.
So, SMARTS parsing takes about 1/9th of the time of the actual graph matching…
Experimenting the new implementations of the core data model interfaces is very welcome! In particular, if the are faster and take less memory… I played with it in the past a bit, but never got performance significantly boosted…
Also, the CDK implementation caught several bugs in my code and did several things more elegantly than mine, which I used to improve my SMARTS patterns.
Any elegance in the CDK code is due to my NCGC colleague, Trung, who did the NCGC implementation which I ported to the CDK