A few days back, Aaron posted a question regarding the use of the tree width of a graph (intuitively, a measure of how tree like a graph is) in a chemical context. The paper that he pointed to was not very informative in terms of chemical applications. The discussion thread then expanded to asking about the utility of this descriptor – could it be used in a QSAR context as a descriptor of molecular structure? Or is it more suitable in a “filtering” scenario, since as Aaron pointed out “Some NP-complete problems become tractable when a graph has bounded treewidth … ” (with graph isomorphism given as an example).
I took a look at the first question – is it a useful descriptor? Yamaguchi et al, seems to indicate that this is a very degenerate descriptor (i.e., different structures give you the same value of the tree width). Luckily, someone had already done the hard work of implementing a variety of algorithms to evaluate tree widths. libtw is a Java library that provides a handy framework to experiment with tree width algorithms. I implemented a simple adapter to convert CDK molecule objects into the graph data structure used by libtw and a driver to process a SMILES file and report the tree width values as well as execution times. While libtw provides a number of tree width algorithms I just used a single one (arbitrarily). The code is available on Github and requires the CDK and libtw jar files to compile and run.
I took a random sample of 10,000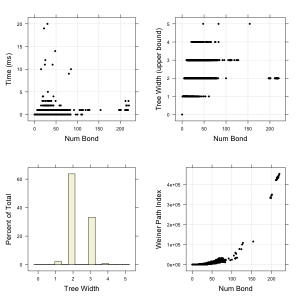 molecules from ChEMBL (also in the Github repository) and evaluated the upper bound of the tree width for each molecule. In addition, I evaluated a few well known topological descriptors for comparison purposes. The four plots summarize the results.
molecules from ChEMBL (also in the Github repository) and evaluated the upper bound of the tree width for each molecule. In addition, I evaluated a few well known topological descriptors for comparison purposes. The four plots summarize the results.
The calculation is certainly very fast, and, surprisingly, doesn’t seem to correlate with molecular size. Apparently, some relatively small molecules take the longest time – but even those are very fast. Unfortunately, the descriptor is indeed degenerate as shown in the top right – a given tree width value shows up for both small and large molecules (the R^2 between number of bonds and tree width is 0.03). The histogram in the lower left indicates that 60% of the molecules had the same value of tree width. In other words, the tree width does not really differentiate bewteen molecular structures (in terms of size or complexity). In contrast, if we consider the Weiner Path index, which has been used extensively in QSAR models, primarily as a measure of branching, we see that it exhibits a much closer relation with molecular size. Other topological measures focusing more specifically on structural complexity such as fragment complexity show similar correlations with molecular size (and with each other).
So in conclusion, I don’t think the tree width is a useful descriptor for modeling purposes.