Earlier today, Emily Wixson posted a question on the CHMINF-L list asking
… if there is any way to count the number of authors of papers with specific keywords in the title by year over a decade …
Since I had some code compiling and databases loading I took a quick stab, using Python and the Entrez services. The query provided by Emily was
1 | (RNA[Title] OR "ribonucleic acid"[Title]) AND ("2009"[Publication Date] : "2009"[Publication Date]) |
The Python code to retrieve all the relevant PubMed ID’s and then process the PubMed entries to extract the article ID, year and number of authors is below. For some reason this query also retrieves articles from before 2001 and articles with no year or zero authors, but we can easily filter those entries out.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 | import urllib, urllib2, sys import xml.etree.ElementTree as ET def chunker(seq, size): return (seq[pos:pos + size] for pos in xrange(0, len(seq), size)) query = '(RNA[Title] OR "ribonucleic acid"[Title]) AND ("2009"[Publication Date] : "2009"[Publication Date])' esearch = 'http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&mindate=2001&maxdate=2010&retmode=xml&retmax=10000000&term=%s' % (query) handle = urllib.urlopen(esearch) data = handle.read() root = ET.fromstring(data) ids = [x.text for x in root.findall("IdList/Id")] print 'Got %d articles' % (len(ids)) for group in chunker(ids, 100): efetch = "http://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?&db=pubmed&retmode=xml&id=%s" % (','.join(group)) handle = urllib.urlopen(efetch) data = handle.read() root = ET.fromstring(data) for article in root.findall("PubmedArticle"): pmid = article.find("MedlineCitation/PMID").text year = article.find("MedlineCitation/Article/Journal/JournalIssue/PubDate/Year") if year is None: year = 'NA' else: year = year.text aulist = article.findall("MedlineCitation/Article/AuthorList/Author") print pmid, year, len(aulist) |
With ID’s, year and author counts in hand, a bit of R lets us visualize the distribution of author counts, over the whole decade and also by individual years.
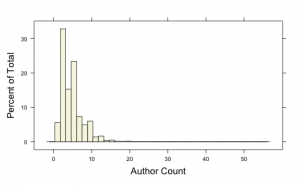
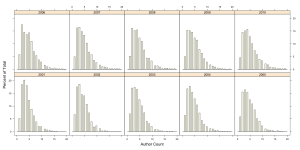
The median author count is 4, There are a number of papers that have more than 15 and some single papers with more than 35 authors on them. If we exclude papers with, say more than 20 authors and view the distribution by year we get the following set of histograms
We can see that over the years, while the median number of authors on a paper is more or less constant at 4 and increases to 5 in 2009 and 2010. But at the same time, the distribution does grow broader over the years, indicating that there is an increasing number of papers with larger author counts.
Anyway, this was a quick hack, and there are probably more rigorous ways to do this (such as using Web of Science – but automating that would be painful).
 I’ve been elected as Chair-Elect of CINF for 2011 so will be switching roles (though I certainly hope to continue contributing to the CINF program in the future).
I’ve been elected as Chair-Elect of CINF for 2011 so will be switching roles (though I certainly hope to continue contributing to the CINF program in the future).