In my previous post I had mentioned that key/value or non-relational data stores could be useful in certain cheminformatics applications. I had started playing around with MongoDB and following Rich’s example, I thought I’d put it through its paces using data from PubChem.
Installing MongoDB was pretty trivial. I downloaded the 64 bit version for OS X, unpacked it and then simply started the server process:
1
| $MONGO_HOME/bin/mongod --dbpath=$HOME/src/mdb/db |
where $HOME/src/mdb/db is the directory in which the database will store the actual data. The simplicity is certainly nice. Next, I needed the Python bindings. With easy_install, this was quite painless. At this point I had everything in hand to start playing with MongoDB.
Getting data
The first step was to get some data from PubChem. This is pretty easy using via their FTP site. I was a bit lazy, so I just made calls to wget, rather than use ftplib. The code below will retrieve the first 80 PubChem SD files and uncompress them into the current directory.
1
2
3
4
5
6
7
8
9
10
11
12
13
| import glob, sys, os, time, random, urllib
def getfiles():
n = 0
nmax = 80
for o in urllib.urlopen('ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/CURRENT-Full/SDF/').read()
o = o.strip().split()[5]
os.system('wget %s/%s' % ('ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/CURRENT-Full/SDF/', o))
os.system('gzip -d %s' % (o))
n += 1
sys.stdout.write('Got n = %d, %s\r' % (n,o))
sys.stdout.flush()
if n == nmax: return |
This gives us a total of 1,641,250 molecules.
Loading data
With the MongoDB instance running, we’re ready to connect and insert records into it. For this test, I simply loop over each molecule in each SD file and create a record consisting of the PubChem CID and all the SD tags for that molecule. In this context a record is simply a Python dict, with the SD tags being the keys and the tag values being the values. Since i know the PubChem CID is unique in this collection I set the special document key “_id” (essentially, the primary key) to the CID. The code to perform this uses the Python bindings to OpenBabel:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
| from openbabel import *
import glob, sys, os
from pymongo import Connection
from pymongo import DESCENDING
def loadDB(recreate = True):
conn = Connection()
db = conn.chem
if 'mol2d' in db.collection_names():
if recreate:
print 'Deleting mol2d collection'
db.drop_collection('mol2d')
else:
print 'mol2d exists. Will not reload data'
return
coll = db.mol2d
obconversion = OBConversion()
obconversion.SetInFormat("sdf")
obmol = OBMol()
n = 0
files = glob.glob("*.sdf")
for f in files:
notatend = obconversion.ReadFile(obmol,f)
while notatend:
doc = {}
sdd = [toPairData(x) for x in obmol.GetData() if x.GetDataType()==PairData]
for entry in sdd:
key = entry.GetAttribute()
value = entry.GetValue()
doc[key] = value
doc['_id'] = obmol.GetTitle()
coll.insert(doc)
obmol = OBMol()
notatend = obconversion.Read(obmol)
n += 1
if n % 100 == 0:
sys.stdout.write('Processed %d\r' % (n))
sys.stdout.flush()
print 'Processed %d molecules' % (n)
coll.create_index([ ('PUBCHEM_HEAVY_ATOM_COUNT', DESCENDING) ])
coll.create_index([ ('PUBCHEM_MOLECULAR_WEIGHT', DESCENDING) ]) |
Note that this example loads each molecule on its own and takes a total of 2015.020 sec. It has been noted that bulk loading (i.e., insert a list of documents, rather than individual documents) can be more efficient. I tried this, loading 1000 molecules at a time. But this time round the load time was 2224.691 sec – certainly not an improvement!
Note that the “_id” key is a “primary key’ and thus queries on this field are extremely fast. MongoDB also supports indexes and the code above implements an index on the PUBCHEM_HEAVY_ATOM_COUNT field.
Queries
The simplest query is to pull up records based on CID. I selected 8000 CIDs randomly and evaluated how long it’d take to pull up the records from the database:
1
2
3
4
5
6
7
8
| from pymongo import Connection
def timeQueryByCID(cids):
conn = Connection()
db = conn.chem
coll = db.mol2d
for cid in cids:
result = coll.find( {'_id' : cid} ).explain() |
The above code takes 2351.95 ms, averaged over 5 runs. This comes out to about 0.3 ms per query. Not bad!
Next, lets look at queries that use the heavy atom count field that we had indexed. For this test I selected 30 heavy atom count values randomly and for each value performed the query. I retrieved the query time as well as the number of hits via explain().
1
2
3
4
5
6
7
8
9
10
11
12
13
| from pymongo import Connection
def timeQueryByHeavyAtom(natom):
conn = Connection()
db = conn.chem
coll = db.mol2d
o = open('time-natom.txt', 'w')
for i in natom:
c = coll.find( {'PUBCHEM_HEAVY_ATOM_COUNT' : i} ).explain()
nresult = c['n']
elapse = c['millis']
o.write('%d\t%d\t%f\n' % (i, nresult, elapse))
o.close() |
A summary of these queries is shown in the graphs below.
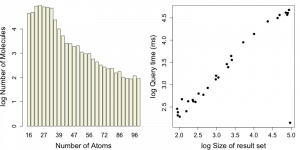
One of the queries is anomalous – there are 93K molecules with 24 heavy atoms, but the query is performed in 139 ms. This might be due to priming while I was testing code.
Some thoughts
One thing that was apparent from the little I’ve played with MongoDB is that it’s extremely easy to use. I’m sure that larger installs (say on a cluster) could be more complex, but for single user apps, setup is really trivial. Furthermore, basic operations like insertion and querying are extremely easy. The idea of being able to dump any type of data (as a document) without worrying whether it will fit into a pre-defined schema is a lot of fun.
However, it’s advantages also seem to be its limitations (though this is not specific to MongoDB). This was also noted in a comment on my previous post. It seems that MongoDB is very efficient for simplistic queries. One of the things that I haven’t properly worked out is whether this type of system makes sense for a molecule-centric database. The primary reason is that molecules can be referred by a variety of identifiers. For example, when searching PubChem, a query by CID is just one of the ways one might pull up data. As a result, any database holding this type of data will likely require multiple indices. So, why not stay with an RDBMS? Furthermore, in my previous post, I had mentioned that a cool feature would be able to dump molecules from arbitrary sources into the DB, without worrying about fields. While very handy when loading data, it does present some complexities at query time. How does one perform a query over all molecules? This can be addressed in multiple ways (registration etc.) but is essentially what must be done in an RDBMS scenario.
Another things that became apparent is the fact that MongoDB and its ilk don’t support JOINs. While the current example doesn’t really highlight this, it is trivial to consider adding say bioassay data and then querying both tables using a JOIN. In contrast, the NoSQL approach is to perform multiple queries and then do the join in your own code. This seems inelegant and a bit painful (at least for the types of applications that I work with).
Finally, one of my interests was to make use of the map/reduce functionality in MongoDB. However, it appears that such queries must be implemented in Javascript. As a result, performing cheminformatics operations (using some other language or external libraries) within map or reduce functions is not currently possible.
But of course, NoSQL DB’s were not designed to replace RDBMS. Both technologies have their place, and I don’t believe that one is better than the other. Just that one might be better suited to a given application than the other.
