In my previous post, I had asked whether we really need AtomContainerSet and other related specialized container classes as opposed to using parametrized List objects. Gilleain had mentioned some issues that might require these specialized classes. But at this point it’s not clear to me what the intended goal for these classes was.
For now, assuming that they are used purely as storage classes, I decided to do some performance testing, comparing AtomContainerSet to ArrayList<IAtomContainer>. Currently these results are based on System.currentTimeMillis() as my YourKit licence has expired. So the results are not as fine grained as I’d like.
To perform the tests I read in 2000 3D structures taken from Pub3D and loaded them into an IAtomContainer[]. I then considered four operations
- Given an empty AtomContainerSet, we simply loop over the 2000-element array and add each molecule to the empty container using addAtomContainer. We do the same thing with an empty ArrayList<IAtomContainer>, but use add. The operation is repeated 100 times and the mean time for the addition of 2000 molecules to the empty containers is taken.
- Randomly access elements of the AtomContainerSet and ArrayList, 50,000 times
- Access each element serially (using for (IAtomContainer x : container) idiom). Averaged 100 times
- Remove 500 molecules by reference, randomly from each container structure
- Remove 500 molecules by index, randomly from each container structure
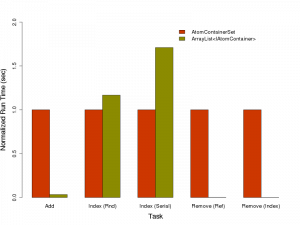
The summary view of the results is shown below. For each pair of bars, the time taken for the operation on the AtomContainerSet is normalized to 1.0 and the time with the ArrayList is appropriately converted.
The raw timings are shown below:
| Operation | AtomContainerSet (sec) | ArrayList (sec) |
| Add | 0.0036 | 0.0001 |
| Indexed Random Access | 0.006 | 0.007 |
| Indexed Serially | 0.007 | 0.012 |
| Remove (by reference) | 68.119 | 0.1 |
| Remove (by index) | 0.695 | 0.0 |
As you can see, the ArrayList is significantly faster in all except random and serial access. In that case, if you consider the raw numbers, the run times are essentially equivalent for random access, though serial access of an array in AtomContainerSet gives it an edge over ArrayList.
So if AtomContainerSet and related classes are purely for storage purposes, they could easily be replaced by ArrayList’s. However the fact that they include support for notifications suggests somebody is using it for more complex scenarios. In either case, a discussion of the design and role of these classes would be useful