Recently, on an email thread I was involved in, Egon mentioned that the CDK hashed fingerprints were probably being penalized by the poor hashing provided by Java’s hashCode method. Essentially, he suspected that the collision rate was high and so that the many bits were being set multiple times by different paths and that a fraction of bits were not being touched.
Recall that the CDK hashed fingerprint determines all topologically unique paths upto a certain length and stores them as strings (composed of atom & bond symbols). Each path is then converted to an int via the hashCode method and this int value is used to seed the Java random number generator. Using this generator a random integer value is obtained which is used as the position in the bit string which will be set to 1 for that specific path..
A quick modification to the CDK Fingerprinter code allowed me to dump out the number of times each position in the bitstring was being set, during the calculation of the fingerprint for a single molecule. Plotting the number of hits at each position allows us to visualize the effectiveness of the hashing mechanism. Given that the path strings being hashed are unique, a collision implies that two different paths are being hashed to the same bit position.
The figure alongside summarizes this for the CDK 1024-bit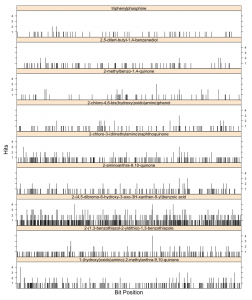 hashed fingerprints on 9 arbitrary molecules. The x-axis represents the bit position and the y-axis on each plot represents the number of times a given position is set to 1 during the calculation. All plots are on the same scale, so we can compare the different molecules (though size effects are not taken into account).
hashed fingerprints on 9 arbitrary molecules. The x-axis represents the bit position and the y-axis on each plot represents the number of times a given position is set to 1 during the calculation. All plots are on the same scale, so we can compare the different molecules (though size effects are not taken into account).
Visually, it appears that the bit positions being set are uniform randomly distributed throughout the length of the fingerprint. However, the number of collisions observed is non-trvial. While for most cases, there doesn’t seem to be a significant number of collisions, the substituted benzoic acid does have a number of bits that are set 4 times and many bits with 2 or more collisions.
The sparsity of triphenyl phosphine can be ascribed to the symmetry of the molecule and the consequent smaller number of unique paths being hashed. However it’s interesting to note that even in such a case, two bit positions see a collision and suggests that the hash function being employed is not that great.
This is a quick hack to get some evidence of hash function quality and its effect on hashed fingerprints. The immediate next step is to look at alternative hash functions. There are also other aspects of the measurement & visualization process that could be tweaked – taking into account molecular size, the actual number of unique paths and converting the plots shown here to some concise numeric representation, allowing us to summarize larger datasets in a single view.
Update – I just realized that the hash function is not the only factor here. The Java random number generator plays an important role. A quick test with the MD5 hash function indicates that we still see collisions (actually, more so than with hashCode), suggesting that the problem may be with how the RNG is being seeded (and the fact that only 48 bits of the seed are used).
Interesting.
I’m relatively new to this fingerprinting stuff, so forgive me if the question is idiotic: would reducing the collisions in a dense bitarray actually help? Surely if you avoid the collisions and fill in the gaps instead, you’d end up with almost every bit set and therefore no useful information (all large molecules would look the same: all bits set).
Humans are notoriously bad at recognizing or generating random number patterns – we always spread things out too much and avoid collisions (e.g. nobody chooses adjacent numbers on their lottery ticket). It may be that what looks to a human like a bad random number or hash code is actually a good one. (Of course, it may also be a bad one).
From one point of view, you’re right. If all bits are being set to one that’s not good – it lowers the resolution, but an easy way out is to use a longer fingerprint. Playing devils advocate – assuming some collision rate is acceptable, how many collisions should we accept?
What is worrying at this point is that different paths are being mapped to the same bit, which ideally, shouldn’t happen.
As far as I can tell this is seems to be due primarily to the RNG rather than the hashing fucntion
An aspect I was a bit more interested in, is to see number of bits that are likely to be set for *a database*. There was some discussion on this a while ago, that the Java hashcode will not use all bits, given a particular chemical dataset; that is, many bits would not be used at all, effectively reducing the size if the fingerprint form 1024, to see 700 or so.
I think this is a generally know problem for many fingerprints, leading to bit selection based on information content, as is done in the MDL fingerprints (Symyx fingerprints?).
So, what I plan to do soon, is run the CDK fingerprints (there are some 4 or 5) against PubChem and calculate the distribution of bits set for the whole of PubChem.
@egon, I’d be interested in seeing that – I’d be surprised if every bit didn’t get set at least once in a data set of PubChem size
[…] my previous post I looked at how many collisions in bit positions were observed when generating hashed fingerprints […]