As part of my work at IU I have been implementing a number of cheminformatics web services. Initially these were SOAP, but I realized that REST interfaces make life much easier. (also see here) As a result, a number of these services have simple REST interfaces. One such service provides molecular descriptor calculations, using the CDK as the backend. Thus by visiting (i.e., making a HTTP GET request) a URL of the form
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/CC(=O)
you get a simple XML document containing a list of URL’s. Each URL represents a specific “resource”. In this context, the resource is the descriptor values for the given molecule. Thus by visiting
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/ org.openscience.cdk.qsar.descriptors.molecular.ALOGPDescriptor/CC(=O)C
one gets another simple XML document that lists the names and values of the AlogP descriptor. In this case, the CDK implementation evaluates AlogP, AlogP2 and molar refractivity – so there are actually three descriptor values. On the other hand something like the molecular weight descriptor gives a single value. To just see the list of available descriptors visit
http://www.chembiogrid.org/cheminfo/rest/desc/descriptors
which gives an XML document containing a series of links. Visiting one of these links gives the “descriptor specification” – information on the vendor, version, reference to a descriptor ontology and so on.
(I should point out that the descriptors available in this service are from a pretty old version of the CDK. I really should update the descriptors to the 1.2.x versions)
Applications
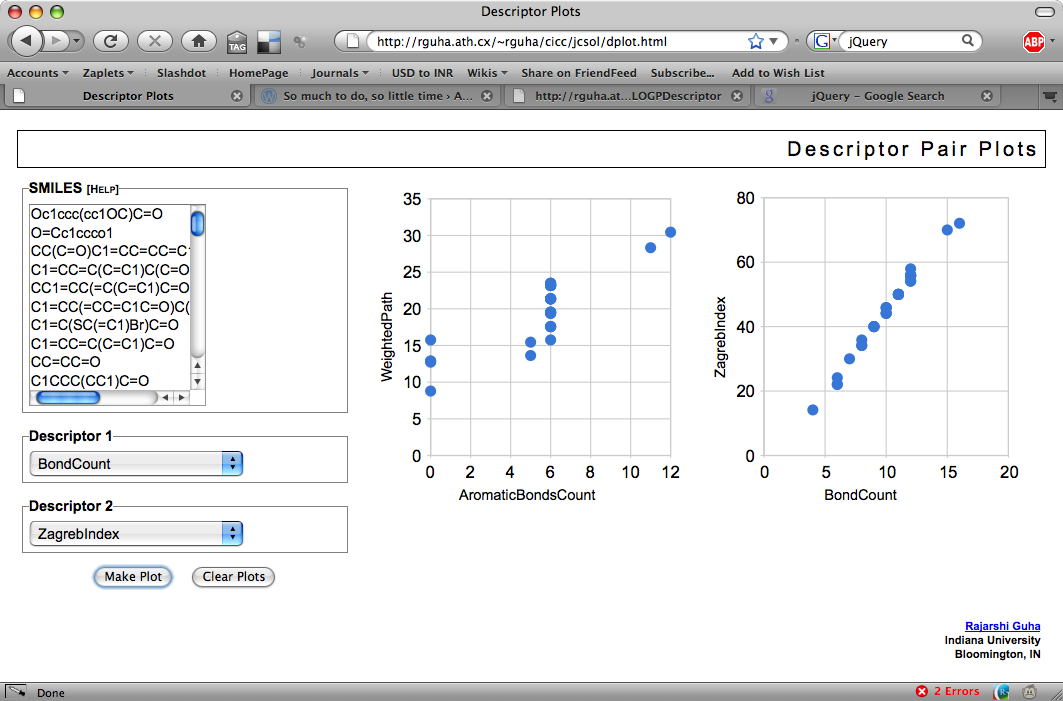
This type of interface makes it easy to whip up various applications. One example is the PCA analysis of compound collections. Another one I put together today based on a conversation with Jean-Claude was a simple application to plot pairs of descriptor values for a collection of SMILES.
The app is pretty simple (and quite slow, since it uses synchronous GET’s to the descriptor service for each SMILES and has to make two calls for each SMILES – hey, it was a quick hack!). Currently, it’s a bit restrictive – if a descriptor calculates multiple values, it will only use the first value. To see how many values a molecular descriptor calculates, see the list here.
With a little more effort one could easily have a pretty nice online descriptor calculation application rivaling a standalone application such as the the CDK descriptor GUI
Also,if you struggle with nice CSS layouts, the CSS Layout Collection is a fantastic resource. And jQuery rocks.
This looks good, but aren’t you going to run into trouble with SMILES containing at triple bond “#”? The “#” character is a fragment identifier in urls. I’ve found it better to base64 encoded the SMILES when passing in a url. Python has a couple of handy functions, urlsafe_b64encode and decodestring that make this easy. There’s also JQuery library for base64 encoding/decoding on the JavaScript side.
Hi Pat, thanks for the pointer. Yes, one user did hit that problem – since he was connecting programmatically, it turned out that urlencode’ing (which replaces the # with %23) it fixed the problem. Ideally, base64 encoding would be best – it’d also avoid the issue of having to deal with the ‘/’ symbol in SMILES, which I got around by playing with the URL – base64 encoding would simplify the code a lot
[…] 11, 2009 by Rajarshi Guha The current version of the REST interface to the CDK descriptors allowed one to access descriptor values for a SMILES string by simply […]
[…] current version of the REST interface to the CDK descriptors allowed one to access descriptor values for a SMILES string by simply […]