Sometime back I described how I was porting the VFLib algorithms to Java, so that we could use it for substructure search, since the current UniversalIsomorphismTester is pretty slow for this task, in general. While I had translated the Ullman algorithm implementation of VFLib and shown that it outperformed the CDK method, it turned out that didn’t work for certain cases such as finding CCC in C1CC1. This was due to a different definition of isomorphism that VFLib used. Instead, I tried to convert the VF2 implementation to Java. The motivation was that it does indeed perform substructure matching as is usually understood in cheminformatics, and was also reported to be extremely fast. Unfortunately, my translation was buggy and I put it on hold.
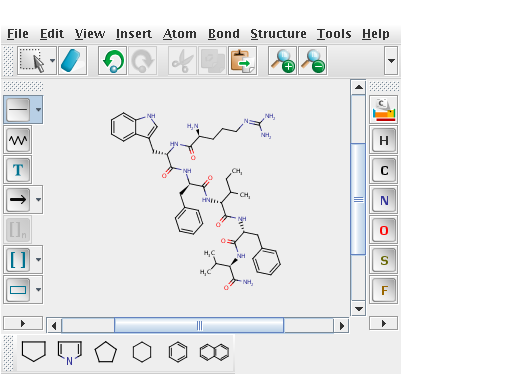
Rich Apodaca took an interest and posted his (initial) port of the VFLib code, based on his own cheminformatics toolkit. Simultaneously, Mark Rijnbeek at the EBI decided to take a look at my code and managed to iron out the bugs, resulting in a Java version of the VF2 and Ullman implementations from VFLib that was based on the CDK for atom and bond matching. He recently sent me some benchmarks, which indicate that VF2 pretty much blows away Ullman for complex queries and is also faster than the current CDK code. He considered the following structure as the query
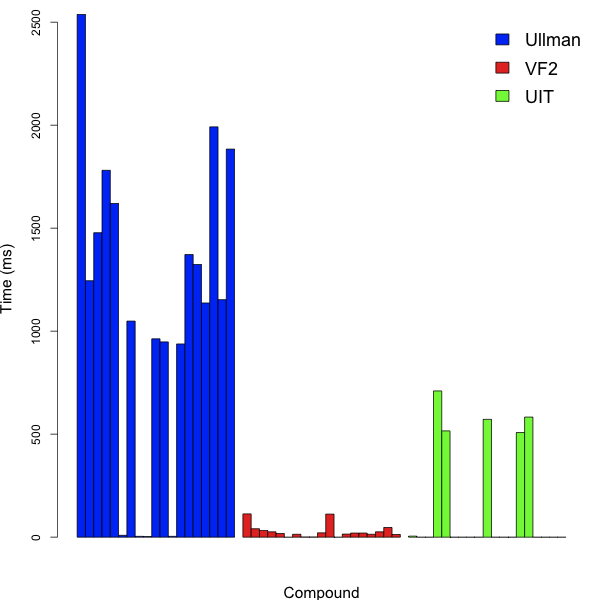
Clearly, this is not a structure that one would commonly use as a query molecule. However, it did occur as a substructure in a number of compounds in the Starlite database and while not necessarily realistic, is a stress test for a subgraph isomorphism algorithm. He then measured the time taken to perform the query against a bunch of compounds, using the Ullman, VF2 and CDK algorithms. The results are summarized below:
For each method, the same target compounds were employed. It should also be noted that the CDK code employs a heuristic filter (if the target has no nitrogens, it can’t match a query containing a nitrogen, etc.). As a result, it breaks out of the search in many cases, resulting in a query time of 0ms. But, impressively, the VF2 code performs very well, even though it is doing a substructure search for each target molecule (i.e., no heuristics to prevent an impossible search from being performed). Now, this is just one benchmark. It will be interesting to see the performance on small query structures – my initial results seem to indicate that VF2 is in general faster than the current CDK code, but not always by a significant amount (mean speedup for a small set of test cases was 17%, with a maximum of 90%). But this needs to be done more rigorously.
But many thanks to Mark for working on this problem – this will be of great benefit to the CDK!
Postscript
Another final issue remains and that is the fact that the VF2 code seems to return the unique set of matches. Thus when matching benzene against benzophenone there are two unique matches (one for each benzene ring). But since each benzene ring can be matched to the query benzene ring in 12 ways, there are a total of 24 non-unique matches. Now, I cannot think of a scenario where one needs the non-unique matches, so it seems that the current form of the VF2 algorithm is good enough. However, I’m probably missing something obvious, so if anybody knows why we might need all the non-unique matches, it would be appreciated.
Non unique matches are useful for calculation of the rmsd to a reference ligand during docking – a ring may have been rotated through 180 degrees with respect to it’s reference, but the resulting docking pose is still a close match to the original. Without checking every permutation of atoms, you’ll not get the lowest possible rmsd value.
I don’t remember ever using non unique matches in any of the 2D cheminformatics tools I have written though.
Thanks Richard – I knew I was missing something!
Rajarshi, great work. Is the benchmarking code available somewhere? Also, how was the matching done (first match or all, for example)?
As a tool to improve performance further, I was thinking it might be useful to create a standard benchmark suite for substructure search. As one of your most recent posts points out, it can be counterproductive to make assumptions about performance without testing and/or profiling.
Rich, I think the code was in the VF2 sources I sent you. In any case, I’ve updated it to a JUnit test – I can send that to you.
I agree that a standard benchmark suite would be good – maybe we can pool our tests and generate a data file with
target query nmatch
(where nmatch = 0 indicates there should be no match)
I had another looks at the VF2 code, especially regarding more complex query molecules like the example pictured above.
The VF2 algorithm tries to make node pairs and for each succesful pair goes down one level deeper in recursion until all pairs are mapped. The graphs are not examined beforehand.
However I thought that some examination might be useful. The pictured query has formula C47 H65 N11 O6. Instead making node pairs unbiased, I sorted the CDK query atom container, putting the least common atoms first in query the atom list . So the six oxygen atoms sit up front, then the eleven nitrogens etc.
That means that the algorithm will start to make node pairs with the least common atoms, and is therefore bound to detect failure quicker. That’s understandable, because mapping C-C pairs is less useful for backtracking than making O-O pairs.
A quick test showed that the sorted query was significantly faster, up to 50%. In the case of C47 H65 N11 O6 about 35% faster.
Mark, great catch! In fact the one of the algorithms in VFLib (either Ulman or VF2) did have a sorting step – which I had not incorporated in the first round, so that I could focus on correctness.
Nice analysis and great results
This paper might be interesting. It has VF implemented in it.
http://www.citeulike.org/user/babakap/article/2937019
Syed, thanks for the pointer. An interesting approach, but I’m not clear as to why it’s an improvement over VFLib