As part of a larger project, I’ve been doing some profiling on various aspects of the CDK, focusing on core cheminformatics operations. I’m using the excellent YourKit profiler to do the tests. They tests are run on a Macbook Pro (2.16GHz) with 1GB RAM, using the latest trunk version of the CDK and JDK 1.5.
The test data is a 1000-molecule subset take from the ZINC collection. The operations I’ve been looking at are
- Ring perception (AllRingsFinder)
- Aromaticity Perception (CDKHueckelAromaticityDetector)
- Atom type perception
- SDF reading (IteratingMDLReader)
- Fingerprinting
- Tanimoto similarity (Tanimoto.calculate())
The test harness simply reads the 1000 molecules one by one and performs the operation in question. For certain tasks which are not atomic in nature, the code does a little more but the timing is measured only for the operation under study. In all cases, things like loading molecules from disk are not measured. The whole process is repeated 10 times and the times reported are the average of the 10 runs. A brief overview of the results:
SDF Reading
Looping over the file of 1000 molecules, with no subsequent operations (such as atom typing or aromaticity detection). Time required = 0.875s
Ring perception
Out of the 1000 molecules, there were 927 molecules with a total of 3090 rings. Time required = 1.027s
Aromaticity perception
Out of the 1000 molecules there were 733 aromatic molecules. Time required = 1.334s
Atom typing
For this test I employed AtomContainerManipulator.percieveAtomTypesAndConfigureAtoms. So the time for this does include the time for reading from the data files (but that’s a one time process). Time required = 1.152s
Fingerprinting
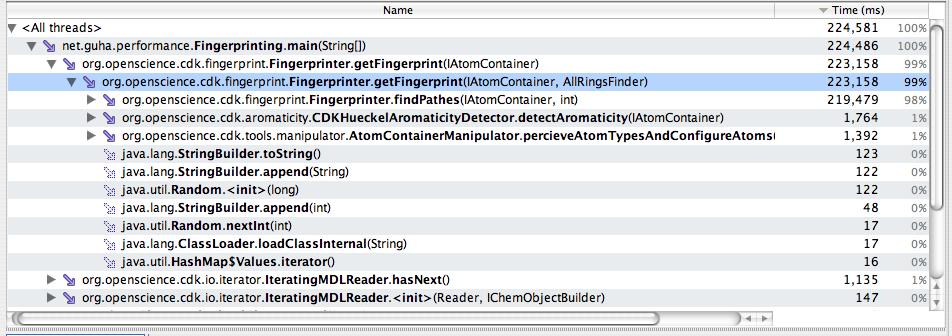
This was tested using the Fingerprinter with default arguments on all 1000 molecules. This method is not “atomic” as something like ring perception since it will perform aromaticity perception and atom typing, but is a common enough cheminformatics task. Time required = 222.456s
The figure below highlights the bottleneck in this method:
Similarity
This is tested by first generating 1000 fingerprints using Fingerprinter and then evaluating unique, pairwise similarity values. The time reported is for the 499500 similarity calculations. Time required = 0.962s
Rajarshi, for the atom typing of Sybyl atom types, I had to introduce a new method in the IAtomTypeMatcher interface, effectively giving two flavors of the same thing. Which of these two did you use:
findMatchingAtomType(IAtomContainer, IAtom)
findMatchingAtomTypes(IAtomContainer)
The first one would manually iterate over the IAtomContainer’s atoms, but also includes some redundant work. So, the second should be faster… Might you compare the two versions, so that maybe we could use that info as example of the amount of performance we gain when we think about performance…?
I used the perceveAtom…() method. I’ll do some tests with these two methods and see what happens
[…] 12, 2008 by Rajarshi Guha In my last post I had reported some timing measurements for various operations. One of them was fingerprinting […]
Did a quick check and if I use findMatchingAtomType(IAtomContainer) directly rather than perceiveAtom…() it takes 0.78s for a 1000 molecules, averaged over 10 runs