As part of an internal project I’ve recently started working with Neo4j for representing and querying relationships between entities (targets, compounds, etc.). What has really caught my attention is the Cypher graph query language – by allowing you to construct queries using graph notation, many tasks that would be complex or tedious in a traditonal RDBMS become much easier.
As an example, I loaded the ChEMBL target hierarchy and the targets as a graph. On it’s own it’s not particularly useful – the real utility arises when other datasets (and datatypes) are linked to the targets. But even at this stage, one can easily ask questions such as
Find all kinase proteins
which is simply a matter of identifying proteins that have a direct path to the Kinase target class.
Assuming you have ChEMBL loaded in to a MySQL database, you can generate a Neo4j graph database containing the targets and classification hierarchy using code from the neo4jexpt repository. Simply compile and run as (appropriately changing host name, user and password)
1 2 3 | $ mvn package $ java -Djdbc.url="jdbc:mysql://host.name/chembl_20?user=USER&password=PASS" \ -jar target/neo4j-ctl-1.0-SNAPSHOT.jar graph.db |
Once complete, you should see a folder named graph.db. Using the Neo4j application you can then explore the graph in your browser by executing Cypher queries. For example, lets get the graph of the entire ChEMBL target classification hierarchy (and ensuring that we don’t include actual proteins)
1 2 | MATCH (n {TargetType:'TargetFamily'})-[r]-(m {TargetType:'TargetFamily'}) RETURN r |
(The various annotations such as TargetType and TargetFamily are based on my code). When visualized we get
Lets get more specific, and extract the kinase portion of the classification hierarchy
1 2 3 4 | MATCH (n {TargetType:'TargetFamily'}), (m {TargetID:'Kinase'}), p = shortestPath( (n)-[:ChildOf*]->(m) ) RETURN p |
Given that we’ve linked the protein themselves to the target classes, we can now ask for all proteins that are kinases
1 2 3 4 | MATCH (m {TargetType:'MolecularTarget'}), (n {TargetID:'Kinase'}), p = shortestPath( (m)-[*]->(n) ) RETURN m |
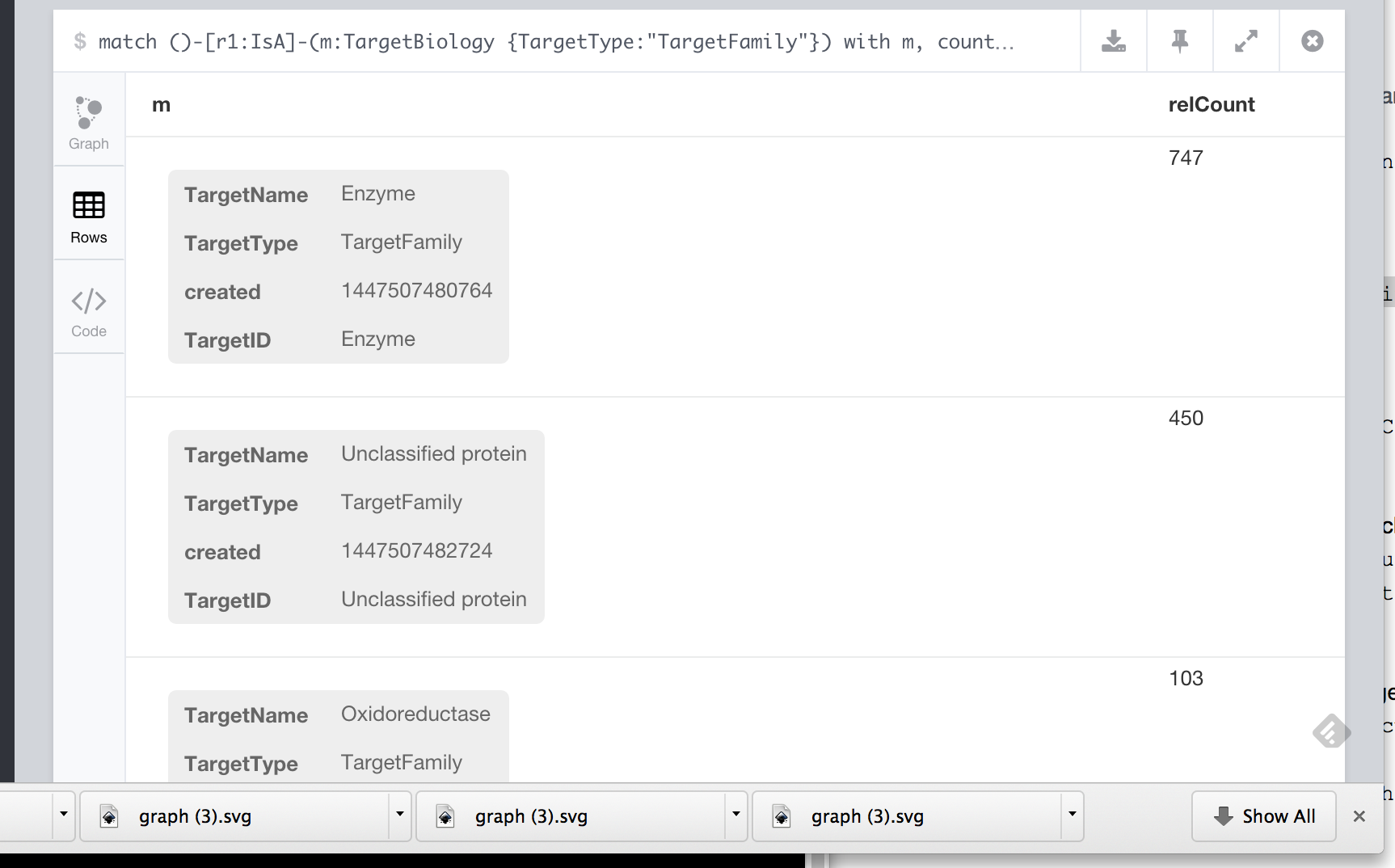
Or identify the target classes that are linked to more than 25 proteins
1 2 3 4 | MATCH ()-[r1:IsA]-(m:TargetBiology {TargetType:"TargetFamily"}) WITH m, COUNT(r1) AS relCount WHERE relCount > 25 RETURN m |
which gives us a table of target classes and counts, part of which is shown below
Overall this seems to be a very powerful platform to integrate data sources and types and effectively query for relationships. The browser based view is useful to practice Cypher and answer questions of the dataset. But a REST API is available as well as other tools such as Gremlin that allow for much more flexible applications and sophisticated queries.